| Company | Pros | Cons |
|---|---|---|
 23andMe |
|
|
 AncestryDNA |
|
|
 FTDNA |
|
|
 MyHeritage |
|
|
If it’s in your budget, Genetic Genie recommends taking a step into the future. Get Whole Genome Sequencing at Nebula Genomics.*
* The above link is an affiliate link. Save $10 at Nebula Genomics with 10OFF until 5/31 and support our efforts.
The Difference Between Genotyping and Whole Genome Sequencing
In this article, we are comparing genotyping companies. If you want the latest and greatest technology, Genetic Genie recommends getting Whole Genome Sequencing. Whole Genome Sequencing allows consumers to get clinical-grade information about all the variants in their genome.
23andMe, AncestryDNA, MyHeritage and 23andMe do not currently offer Whole Genome Sequencing tests. Instead, they use genotyping arrays that give you around 0.02% of your DNA. Data scientists try their best pick the most important variants, but a lot of important and/or currently unclassified health-related variants will be completely missing from raw DNA data files. And a lot of health-related variants (especially insertion/deletion variants like BRCA) are often not correctly called by these genotyping arrays. So when using third-party tools to analyze your raw data, you have to be aware of these limitations. Errors in interpretation aren’t necessarily the fault of third-parties. If third-parties don’t filter out every miscall from these genotyping arrays (which is very hard), you may end up with some erroneous information.
While genotyping had its limitations, these tests are generally more useful than Whole Genome Sequencing for general ancestry purposes. However if you are male, technical, and are into Genetic Geneology, you can use a service like YFull for a very advanced analysis of your Y-DNA from Whole Genome Sequencing data.
Raw Data SNP Comparison
Chip versions are included for the Total SNPs. Other information is based on the latest chip versions when this article was published. This table describes the basic raw data offerings without add-on tests.
(table scrollable on mobile device)
| 23andMe | AncestryDNA | FamilyTreeDNA (FTDNA) | My Heritage DNA | |
|---|---|---|---|---|
| Total SNPs | v2: ~555,000 v3: ~900,000 v4: ~570,000 v5: ~640,000 | v1: ~700,000 v2: ~664,000 | ~702,000 |
v1: ~708,000 v2: ~720,000 |
| ClinVar SNPs | ~33,000 | ~76,000 | ~19,000 | ~20,000 |
| Drug Response | 215 | 353 | 103 | 104 |
| X SNPs | 16531 | 24887 | none | 17,892 |
| Y SNPs | 3,733 | 1,803 | none without add-ons | 482 |
| mtDNA SNPs | 4301 | 224 | none without add-ons | none |
| Autosomal SNPs | 614,005 | 637,469 | 702,433 | 702,430 |
SNP data computed internally by Genetic Genie. ClinVar data based on 5/7/2020 release.
23andMe has several versions of their chips. Each chip has a different number of SNPs (Single Nuceleotide Polymorphisms). 23andMe’s V2 chip from 2009 has about 555,000 SNPs, the v3 chip from 2010 has about 900,000 SNPs, the v4 chip from 2013 has about 570,000 SNPs, and their current v5 chip (2017 to present) has about 640,000 SNPs. AncestryDNA has about 700,000 SNPs on their v1 chip and about 664,000 on their current v2 chip. MyHeritage’s autsosomal v1 chip has about 708,000 variants and their v2 chip has about 720,000 SNPs. FamilyTreeDNA (FTDNA) has about 702,000 SNPs. However, FTDNA doesn’t come with Y-DNA or mtDNA and adding a full mtDNA sequence or extra Y-DNA information can be costly. However, you can upload your full mtDNA sequence FASTA file to third parties like MITOMASTER for haplogroup prediction.
In terms of clinical SNPs, 23andMe currently has approximately 33,000 SNPs in ClinVar in their v5 chip, FamilyTreeDNA (FTDNA) has about 19,000 ClinVar SNPs, My Heritage DNA has around 20,000 ClinVar SNPs and AncestryDNA has more than twice as many ClinVar SNPs at around 76,000.
Clinically-relevant SNPs may not be a huge deal if your primary interest is in ancestry. But if you want to take a peek at health related variants with third party apps, it’s important to know what’s in the product you are getting.
FTDNA and MyHeritage DNA are tied for the most autosmal SNPS at about 702,430. In terms of other non-autosomal SNPs, AncestryDNA has the most X SNPs at around 24,887. 23andMe has the most mtDNA SNPs at 4,301.
Getting more SNPs with merging and imputation
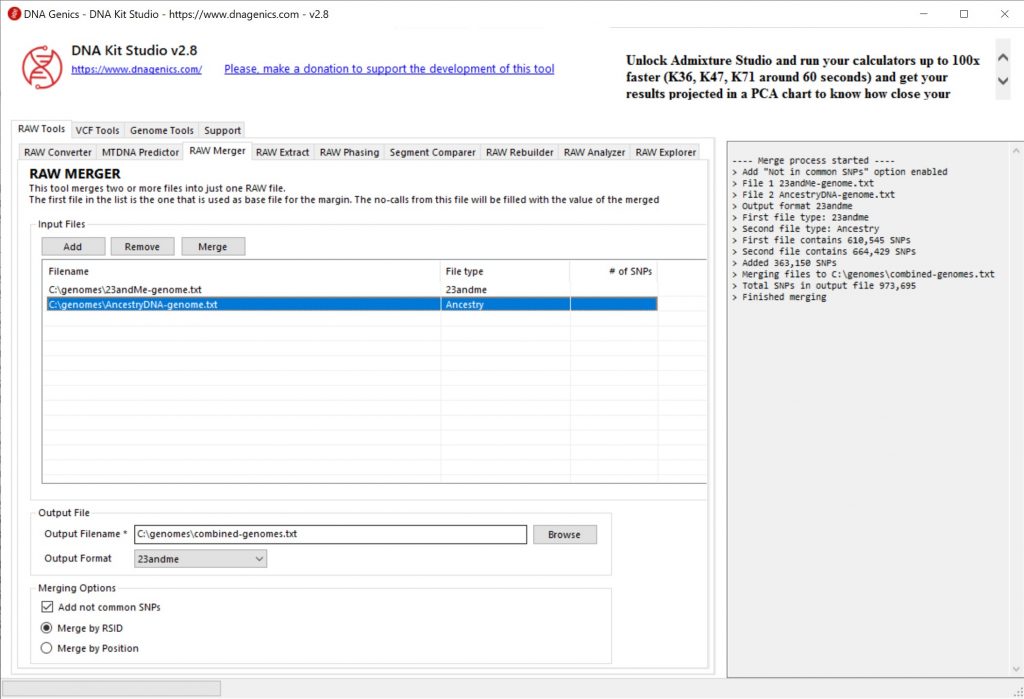
To merge files, you can use the Raw Merger option in the Windows Application DNA Kit Studio. Of course, there are countless technical ways to accomplish this on the Linux and Mac command line, but this article is designed to be newbie friendly. You can merge multiple AncestryDNA files, multiple 23andMe files, a mix of 23andMe and AncestryDNA files, etc. If you’ve been tested multiple times on multiple chips, merging files will give you a lot more SNPs in a single raw DNA data file.
Combined files can be useful for both health analysis and genetic genealogy. Combining kits can also increase accuracy in your match information when using the online service GEDmatch.
We’re not going to get into what imputation is or how it works in this article, but imputation is an easy (and magical) way to get some extra SNPs. You can upload your 23andMe formatted file Michigan Imputation Server to get more variants.
This process is fairly accurate but is also error-prone. This is probably fine for ancestry purposes but should not be trusted if you are looking at health variants. It still may be fun to get a few more health related variants out of curiosity, but I definitely wouldn’t put any faith into such data if you are planning to use imputed data for health analysis.
The Winner is AncestryDNA
We find AncestryDNA to be the best DNA raw data test kit for both ancestry and health. Not only does 23andMe have significantly less health variants than AncestryDNA, but many variants have custom proprietary identifiers (often important insertion/deletion indel variants) that are miscalled. With some effort, these proprietary variant identifiers can be 100% successfully decoded into their non-proprietary forms, but why did they bother? But 23andMe does deserve some credit as they provide the most Y SNPs and mtDNA variants in their raw data file. 23andMe provides raw data at an affordable price that is well-rounded for genetic geneoological purposes, especially if mtDNA data and Y SNPs are important to you.
AncestryDNA is not perfect either. It has plenty of miscalls for health-related variants. But it is still the best and provides well-rounded consumer genotyping data for both ancestry and health. However, 23andMe is significantly better for mitochondrial DNA analysis and has a lot more Y SNPs, so combining raw data from 23andMe and AncestryDNA using DNA Kit Studio may be the best bargain.
FamlyTreeDNA (FTDNA) is trusted by amateur and professional genealogists but is not good for health analysis. If genealogy is your thing, you might want to consider FamilyTreeDNA. FamilyTreeDNA can do add-on tests such as sequencing of the Y chromosome and mitochondrial DNA. You can receive your full mtDNA in FASTA format and your Y Sequencing in BAM format for upload to services like Y Full. However, it’s pricey: Currently $159 for your mtDNA sequence and $449 for the Big Y-700. These add-ons are not standard genotyping and therefore are separate from their DNA raw data genotyping file. At this point at time it’s more affordable to get a Whole Genome Sequence (WGS) test and extract your Y DNA and mtDNA yourself. This takes a little bit of technical skill and plenty of hard drive space, but we think it’s worth it.
If you prefer FamilyTreeDNA’s data for your uses but want a good amount of X SNPs and a few Y SNPs, MyHeritage DNA may be the best choice. Like FTDNA, MyHeritage DNA’s data is good for ancestry use but not health analysis. However, MyHeritage DNA does not have add-ons for a full mtDNA sequence or Y chromosome sequencing.
There is no one-size-fits-all recommendation for everyone, but we think AncestryDNA’s raw data is best choice for the majority of people — especially if one has an interest in both ancestry and health.
The best DNA raw data genotyping tests in order from best to worst are:
- AncestryDNA (Starting at $99)
- 23andMe (Starting at $99)
- FamilyTreeDNA (FTDNA) (Starting at $79)
- MyHeritage DNA ($79)
AncestryDNA is the best genotyping raw data for uploading to health raw data interpretation/analysis services such as Genetic Genie, GenVue Discovery, Promethease, StrateGene and Xcode Life, and others. With 637,469 autosomal SNPs, AncestryDNA raw data is also a good option for services like GEDmatch and DNA.Land.
Whole Genome Sequencing (WGS) Is Better and Is the Future
If you want data with accuracy and very few miscalls, check out companies like Nebula Genomics or Dante Labs. I’ve done Whole Genome Sequencing through Dante Labs myself, but I’m inclined to say that Nebula has a better science team and is more reputable.
Nebula Genomics has in-depth Whole Genome Sequencing ancestry tools. They can look at your Y-DNA, your mtDNA and have formed a partnership with Family Tree DNA (FTDNA) so you can access the world’s largest Y-DNA and mtDNA databases. It’s a couple hundred extra bucks, but I think it’s worth the investment. I’ve sent in my Nebula kit and am waiting for results.
The data I have received from Dante Labs was very good, but their customer service is par at best. Dante is more focused on health-related variants, but customers have reported errors in their reports. Dante has been trying (they even created their own sequencing center), but they have struggled with customer service, timeliness (though they have a much faster turnaround since they built their own sequencing center), and getting the data right.

Has this changed as of 2024? I have a bunch of MyHeritage raw data files for my family if you want some data for updates. MyHeritage has a health product now that is a one click upgrade, so they might’ve change their set. Also it’s perpetually on sale for $40-$35 now, so it’s definitely the cheapest set.
Thank you for this service, it has been very helpful, one of the very few that can handle WGS files, which I appreciate. Donated a while back.
Hi! thanks for the comparison!
But… could you compare them with sequencing.com and add it to the RAW data table?
Seems it could be better
I was trying to upload my raw data from My Heritage, the system says invalid file. Any ways to fix this? Thank you
You can still do the other two (methylation and detox panels) I think my heritage doesn’t have enough info/ isn’t int he right format for Vue
Hi Kyle,
Do I understand correctly that 23andme Genealogy and Genealogy+Health kits are actually genotyped on the same chip, and provide the same raw DNA information? Only the feedback they give you on health predispositions differs, right?
In that case, one might as well purchase the Genealogy only kit, and use Genetic Genie to recover health information. (and donate the price difference!)
Am I correct?
You are correct. The same raw data is provided no matter what kit you choose.
I have the raw data downloaded and it’s not clear at all where do go once I have it.
Geneticgenie it’s simply amazing.
Fast, private, efficient, complete and easy.
One of the very few instruments to analyze wgs
Thank you!